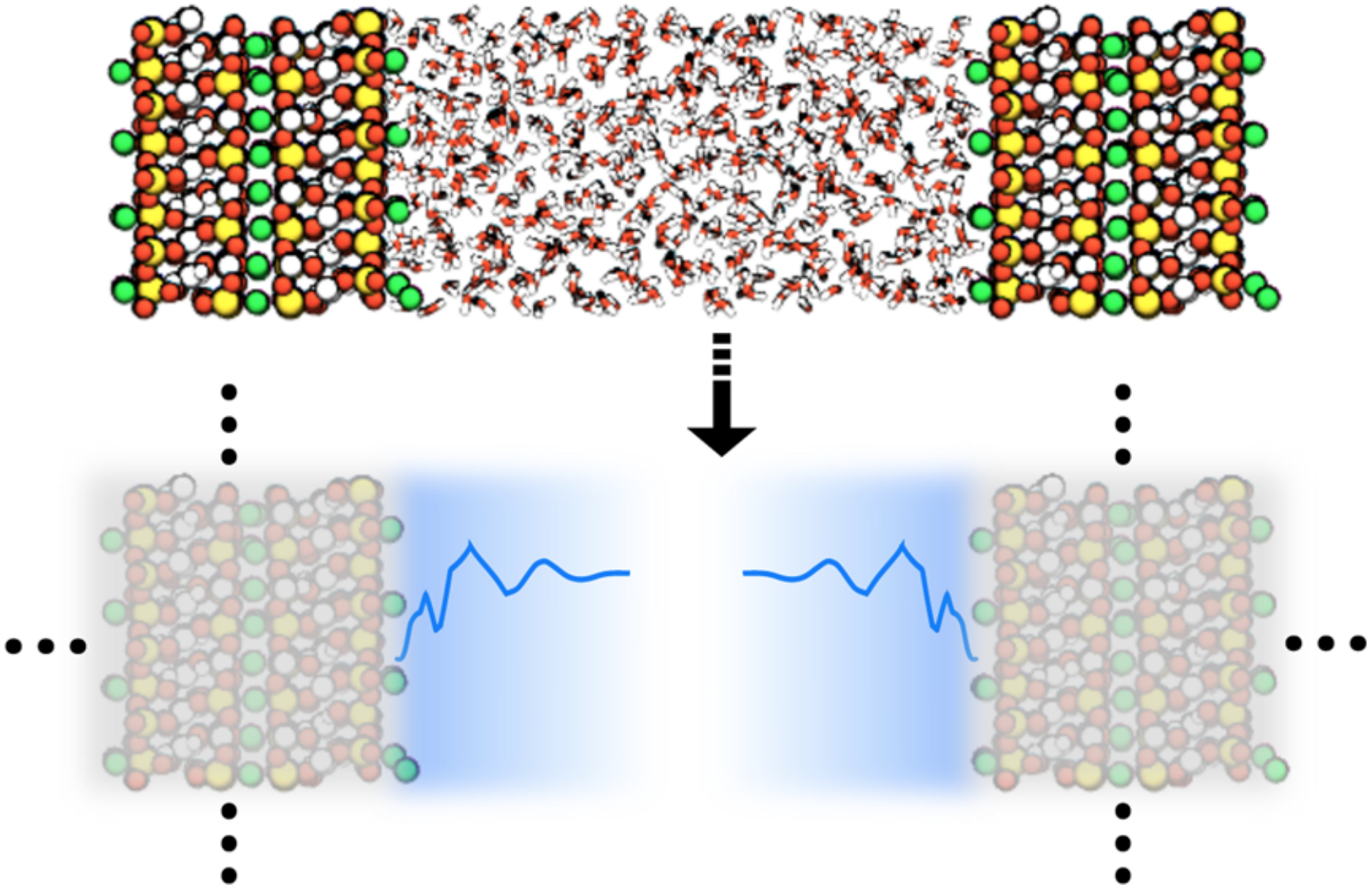
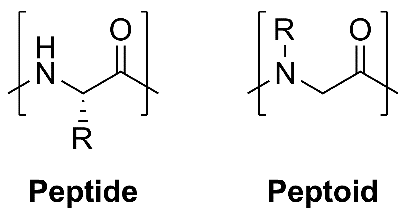
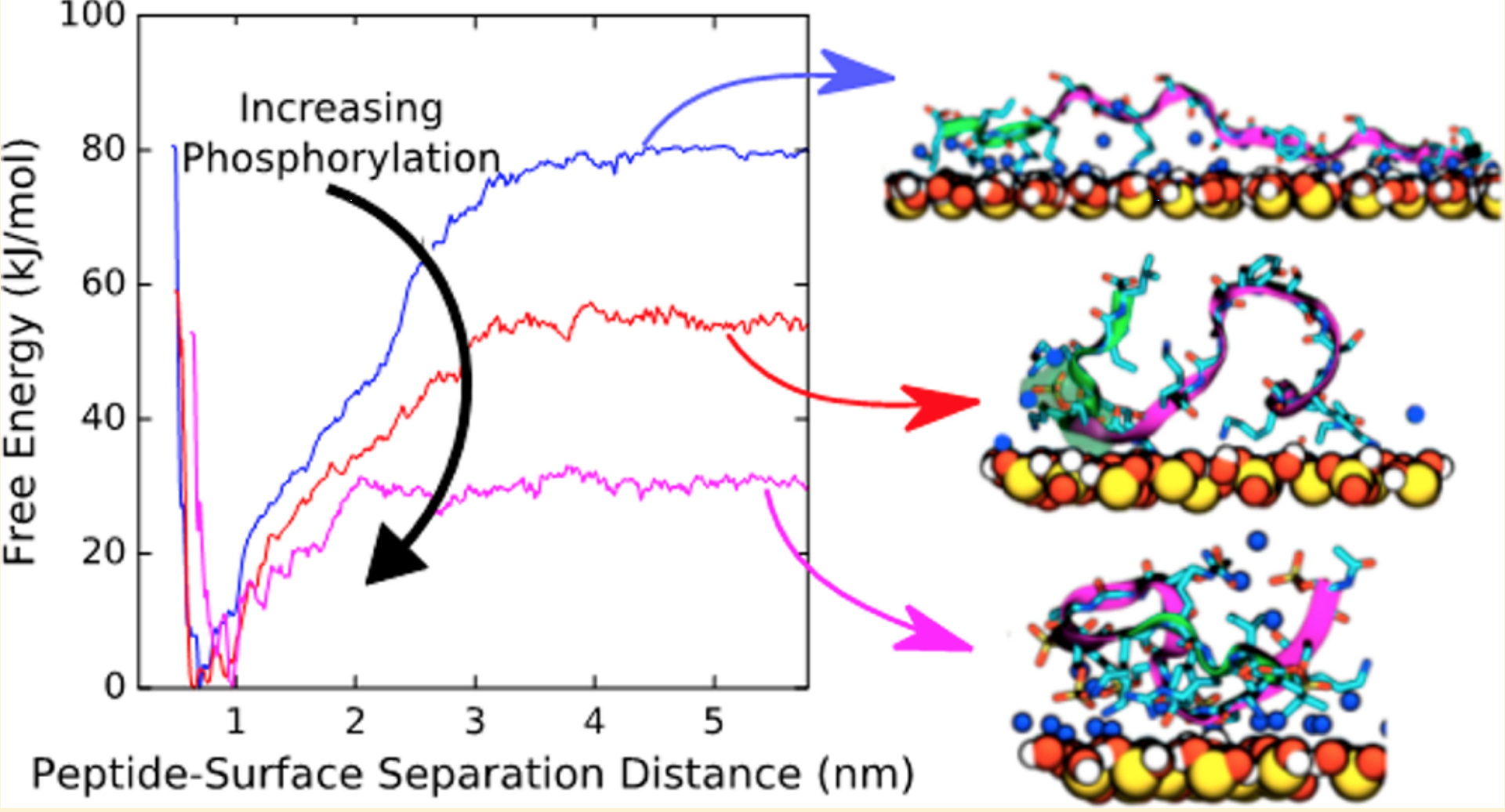
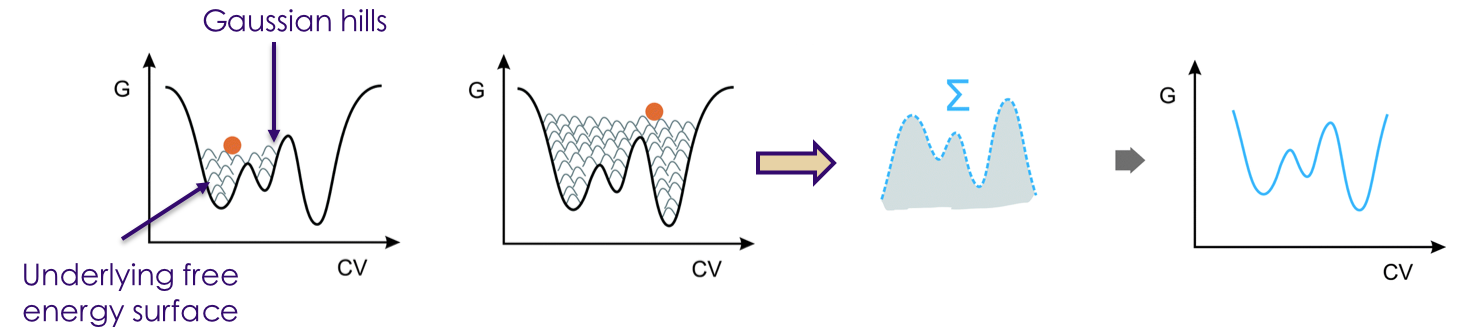
Research Synopsis - Proteins, polymers, and nanoparticles self-assemble in complex ways
into structures like fibrils that cause Alzheimer's disease, mesh structures for use as hydrogels,
and nanotubes for catalysis, respectively. These assemblies occur in solution and at interfaces.
By modeling assembly processes using physics-based simulations (all-atom molecular
dynamics simulations and advanced sampling methods), I am interested in providing mechanistic
insights into these processes. Further, I am also interested in developing methods
and protocols to improve simulations of these systems.
Training - I am a Ph.D. candidate in the Department of Chemical Engineering at the University of Washington (UW), where I work with two fantastic advisors -
Jagjeet and Janice Bindra Endowed Associate Professor
Jim Pfaendtner at UW
and Senior Scientist Christopher J. Mundy
at the Pacific Northwest National Laboratory - and a wonderful research group.
My work is supported by the Materials Synthesis and Simulation Across Scales (MS3) Initiative at PNNL.
Right before heading UW in 2014, I completed my undergraduate degree in Chemical Engineering from
Birla Institute of Technology & Science, Pilani, India
Link to CV
Updates -
Mar 2018 : Successfully passed my Candidacy Exam!
Feb 2018 : GeekWire's coverage of the NW IMPACT association between UW and PNNL
article
Jan 2018 : Recognition by UW's Society of Women Engineers at their annual banquet
picture
Nov 2017 : My work is featured in the video for the MS3 Initiative at PNNL here
Aug 2017 : Made it to the cover of the Journal of Physical Chemistry C! here